Phosphorylation of Cdc6 by CyclinA-Cdk2
Background
Progression through the cell cycle is mediated by cyclin-dependent kinases (Cdk), which regulate a wide range of substrates by phosphorylation. The importance of tight control of Cdk activity is reflected by the wide variety of mechanisms available to regulate these kinases, including association with cyclins, phosphorylation and inhibitor binding (Brown et al., 1999) (Harper and Adams, 2001) (Morris et al., 2002) (Grimmler et al., 2007). One of the substrates of the Cyclin A-Cdk2 complex is the Cdc6 protein, a component of the prereplication complex. As part of the prereplication complex, Cdc6 is involved in loading of the minichromosome maintenance (Mcm) complex onto chromatin, which induces DNA replication. It is of vital importance that there is only a single round of DNA replication per cell cycle. One mechanism that prevents re-initiation is removal of Cdc6 from the nucleus. Translocation of Cdc6 to the cytoplasm is induced by phosphorylation of Cdc6 by the Cyclin A-Cdk2 complex. Activation of Cdk2 for phosphorylation of Cdc6 requires association with Cyclin A and phosphorylation, which is itself dependent on binding of Cyclin A (Petersen et al., 1999) (Cook et al., 2002) (Woo and Poon, 2003) (Cheng et al., 2006). The molecular mechanisms that mediate activation of Cdk2 and result in phosphorylation of Cdc6 are further described below.
Molecular mechanisms
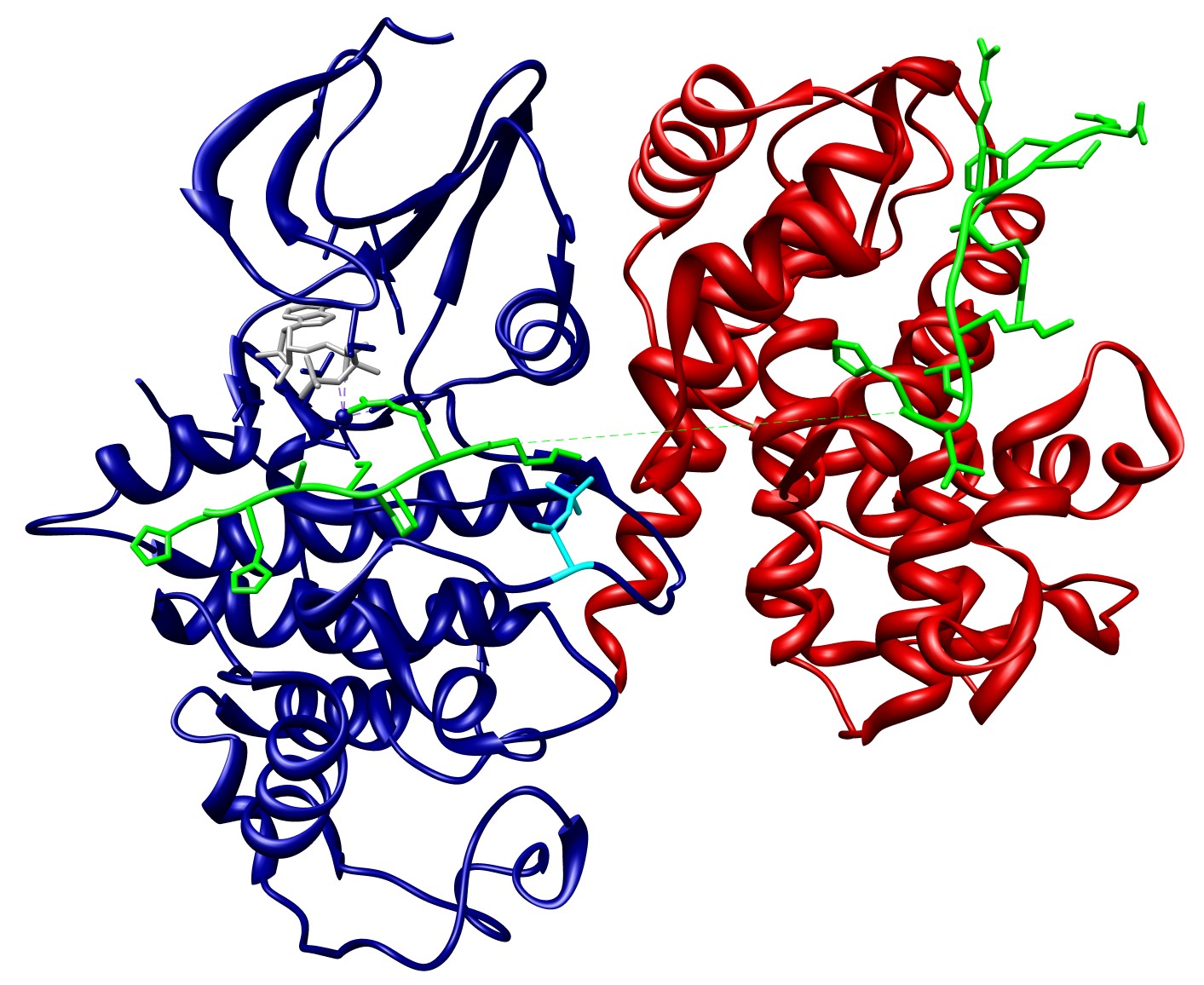
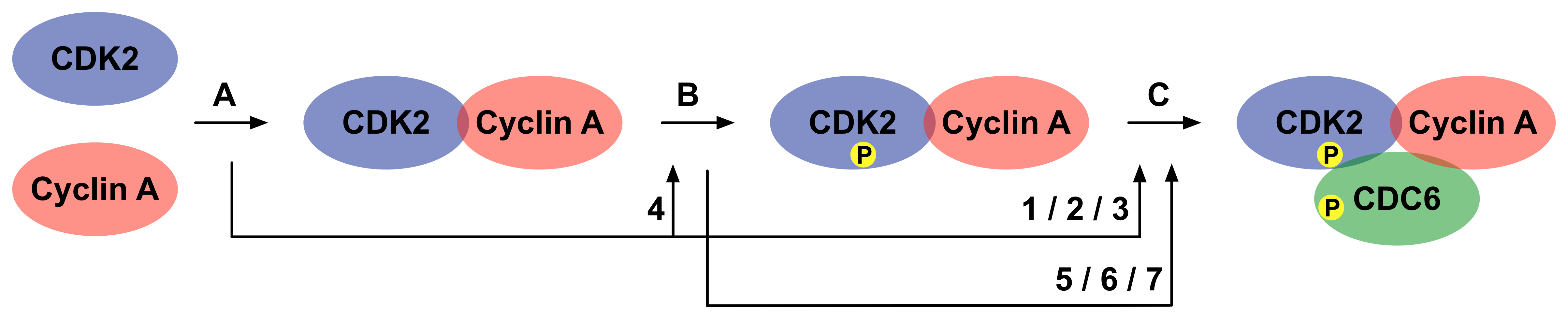
Phosphorylation of Cdc6 by the Cyclin A-Cdk2 complex requires hierarchical build-up of the Cyclin A-Cdk2-Cdc6 complex. Sequential interactions occur in an ordered manner and cooperate to mediate the assembly of the active Cyclin A-Cdk2 complex, and ultimately phosphorylation of Cdc6 (Figure 7). These interactions are interdependent and affect each other through allostery, where PTM or binding of an effector to one site modulates the binding or catalytic properties of a distinct site, and pre-assembly, where the strength of an interaction depends on pre-formation of a complex (Whitty, 2008) (Fenton, 2008).
Binding of Cyclin A to Cdk2 induces allosteric effects in Cdk2. First, it results in structural reorganisation of the ATP binding site of Cdk2 into a conformation optimal for catalysis. Second, it results in structural reorganisation of the T-loop of Cdk2, leading to exposure of the T160 residue, which is now accessible for phosphorylation. In addition, pre-assembly of Cyclin A and Cdk2 results in the formation of a bipartite binding site for Cdc6, with one site on Cyclin A (the substrate recruitment site that interacts with the cyclin recognition site in Cdc6 (LIG_CYCLIN_1)), and one site on Cdk2 (the activation segment). Subsequent phosphorylation of T160 in Cdk2 by Cdk7 (Cak1) results in a conformational change that makes a binding pocket accessible for the proline in the P+1 position on the substrate. These cooperative interactions mediate assembly of an active Cyclin A-Cdk2 complex in a context-dependent manner, provide substrate specificity to the kinase and allow robust regulation of Cdc6 function (Figure 7) (Cheng et al., 2006).
Current representation in Bioinformatics
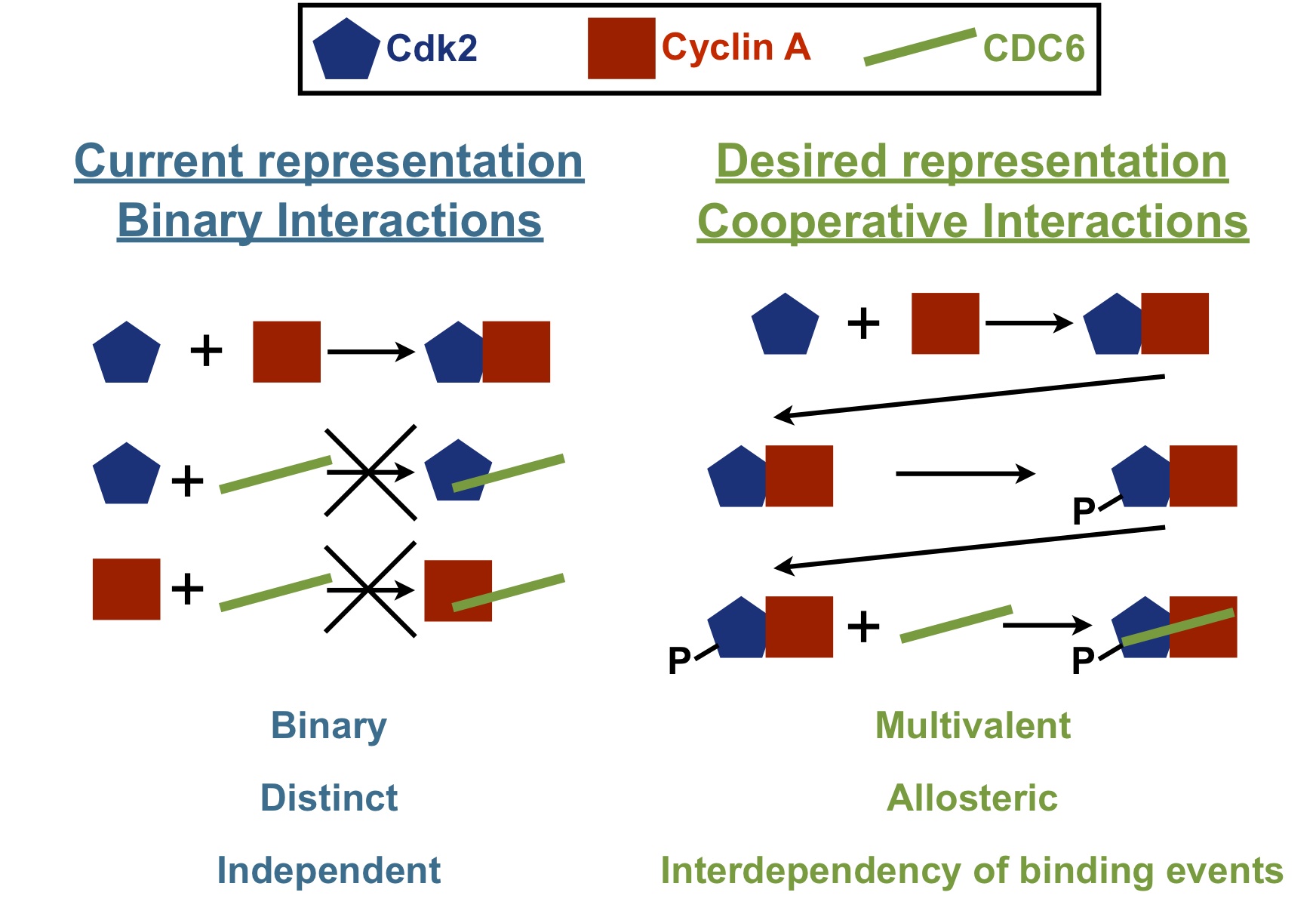
Despite the importance of cooperative interactions in regulating the function of biological systems, they are currently lacking from widely used resources, for instance interaction databases such as IntAct and the Database of Interacting Proteins (DIP), but also from pathway databases like Reactome and the Kyoto Encyclopedia of Genes and Genomes (KEGG) resource. Although these resources provide a large amount of useful and detailed data, the true biological complexity of the interactions and processes they describe can in many cases not be inferred, due to the binary approach of capturing and representing the data, and the lack of annotating interdependencies between different events. In some cases, annotating interaction data in this way can even be misleading. The underlying reason for this is the lack of a computer-readable standard data representation format that is able to capture these interdependencies.
The discrepancies between the complexity of in vivo biological interactions and processes and their in silico representation are illustrated here by means of the example that was described previously, the phosphorylation of Cdc6 by the Cycin A-Cdk2 complex. From the previous description it is clear that activation of Cdk2 and substrate recruitment depend on a sequence of interdependent interactions, including post-translational modification, which allows tight, context-dependent regulation of Cdk2 activity and Cdc6 function. Looking at these interactions in different resources does not allow to infer the complexity of this system as it is in vivo (Figure 8). Representing the interactions between the different components of the complex as binary interactions without annotating interdependencies between them oversimplifies the system, and can even lead to misinterpretation. Therefor, it would be more desirable if the cooperative nature of molecular interactions is captured in a standardised manner (Figure 9).
Representation in PSI-MI using new CV terms
Using the new controlled vocabulary terms that were defined to enable annotation of cooperative interactions using the PSI-MI standard data exchange format, the interdependencies between the interactions that mediate phosphorylation of Cdc6 by the Cyclin A-Cdk2 complex can be captured in significant detail and in a semi-structured manner, allowing to reflect the complexity involved in regulation of such molecular systems.
XML and HTML files
This HTML output shows the phosphorylation of Cdc6 by the Cyclin A-Cdk2 complex, as annotated in a PSI-MI XML file. What is shown is a list of the interactors that participate in the different interactions, and the interactions that result in the assembly of the Cyclin A-Cdk2-Cdc6 complex, as well as the phosphorylation of Cdc6 in this complex. For each interaction, the name, interaction type and participants are described. If an interaction has a cooperative effect on a subsequent interactions, this effect is fully annotated using interaction attributes, as explained previously. If an interaction has more than one cooperative effect, it has to be repeated for each effect. The interactions and cooperative effects are also listed below in more detail.
Interactions
To annotate the phosphorylation of Cdc6 by the Cyclin A-Cdk2 complex in a PSI-MI XML file, 3 interactions are described (Figure 10):
A: Binding of Cyclin A to Cdk2 (named 'CyclinA-Cdk2' in the XML/HTML file).
B: Phosphorylation of Cdk2 on T160 (named 'CyclinA_Cdk2-Cdk7' in the XML/HTML file).
C: Phosphorylation of Cdc6 by the Cyclin A-Cdk2 complex, phosphorylated on Cdk2 (named 'CyclinA_pCdk2-Cdc6' in the XML/HTML file).
Cooperative effects
If one of these interactions has a cooperative effect on a subsequent interaction, this effect is annotated in the attribute list of the interaction. If an interaction has more than one cooperative effect, the complete interaction element has to be repeated for each of these cooperative effects (Figure 10). The cooperative effects are:
1: Binding of Cyclin A to Cdk2 results in the formation of a bipartite binding site for Cdc6, with one site on Cyclin A and one site on Cdk2. Again, the cooperative effect is positive, however the underlying mechanism is pre-assembly, which mediates configurational pre-organization of the interacting components, thereby reducing the entropic costs of their interactions. The interaction that is affected is interaction C (Cheng et al., 2006).
2: Binding of Cyclin A to Cdk2 results in structural reorganisation of the ATP binding site of Cdk2 into a conformation optimal for catalysis. The mechanism underlying this positive effect is allostery, and the interaction that is affected is interaction C. The allosteric molecule is Cdk2, while the allosteric effector is Cyclin A. Since binding of Cyclin A affects the catalysis of Cdk2, this is an allosteric v-type response, mediated by a change in local structure. Since Cyclin A and Cdc6 are not chemically identical, heterotropic allostery occurs (Jeffrey et al., 1995) (Stevenson et al., 2002).
3: Binding of Cyclin A to Cdk2 also has a positive allosteric k-type response on interaction C, due to structural rearrangements that facilitate binding of the substrate (Jeffrey et al., 1995) (Stevenson et al., 2002).
4: Binding of Cyclin A to Cdk2 results in structural reorganisation of the T-loop of Cdk2, leading to exposure of the T160 residue, which is now accessible for phosphorylation. Again, this effect is positive and mediated by allostery, and again the allosteric molecule is Cdk2, while the allosteric effector is Cyclin A. However, now the affected interaction is interaction B, and since it is the binding properties of Cdk2 for its kinase that is altered, this is an allosteric k-type response, also mediated by a change in local structure. The type of allostery is again heterotropic (Jeffrey et al., 1995).
5: Phosphorylation of T160 in Cdk2 by Cdk7 (Cak1) results in a conformational change that makes a binding pocket accessible for the proline in the P+1 position on the substrate. This positive effect is mediated by allostery, and the interaction that is affected is interaction C. Instead of an allosteric effector, the allosteric response, which is an allosteric k-type response, is induced by an allosteric post-translational modification, the phosphorylation of T160 in Cdk2. The allosteric molecule is Cdk2. The allosteric mechanism is again a change in structure, while the type of allostery is again heterotropic (Russo et al., 1996) (Stevenson et al., 2002).
6: Phosphorylation of T160 in Cdk2 by Cdk7 (Cak1) also positively affects the catalytic rate of interaction C by an allosteric mechanism (v-type response) (Russo et al., 1996) (Stevenson et al., 2002).
7: The phosphate moiety on T160 in Cdk2 directly contacts Cdc6, and thereby alters the physicochemical compatibility of Cdk2 for Cdc6 in a positive manner. This means that interaction C is affected by pre-assembly of Cdk2 and the phosphate group on its T160 residue (Brown et al., 1999).
Other examples
Links to relevant publications and data resources can be found in the HTML files.
Cooperative recruitment of p27 to SCF ubiquitin ligase by Skp2 and Cks1.
Pre-assembly of Skp2 and Cks1 results in the formation of a composite binding site that is required to bind the Cyclin-Cdk inhibitor p27. This interaction recruits p27 to the SCF ubiquitin ligase by which it is marked for proteasomal degradation.
Allosteric regulation of dimethylated histone H3 binding to Pygo1 by Bcl9.
Binding of Bcl9 HD1 domain to the PHD-finger of Pygo1 promotes binding of dimethylated histone H3 to a distinct site on Pygo1 through an allosteric mechanism.
Cooperative binding of Plcg1 to LAT.
Recruitment of Plcg1 to LAT-nucleated T cell signalling complexes depends on multivalent binding through its SH2 (which binds to phosphotyrosine motifs in LAT) and PH (which binds to membrane-embedded phosphoinositides) domains. These discrete binding events enhance each other through configurational pre-organisation.
Allosteric phosphorylation-dependent binding of CheY to FliM.
Phosphorylation of CheY at one site stabilises its active conformation, which has increased affinity for the FliM protein binding at a distinct site.
Hydroxylation-induced binding of Hif1a to pVHL.
Enzyme-catalysed proline hydroxylation of pVHL-binding motifs in Hif1a induce binding to pVHL, which targets Hif1a for proteasomal degradation under normoxic conditions.
Phospho-dependent blocking of Stg-Dlg4 binding.
Phosphorylation of the PDZ-binding motif in Stargazin (Stg) abrogates binding to the PDZ domains of Dlg4.
Blocking of the Mkl1 NLS by G actin.
Binding of monomeric G-actin to the RPEL motifs in Mkl1 hides the overlapping nuclear localisation signal (NLS), thereby preventing binding of Mkl1 to the nuclear import machinery and retaining this transcription factor in the cytoplasm.




